NDEx Release Notes - Q4-2024 Update (v2.5.8)
November 12th, 2024
Overview
The Q4-2024 Update is a maintenance release and does not include any new features or API changes. Improvements in this release:
- The NDEx server has been upgraded to use Keycloack for user's sign in, registration and password recovery. If you have an existing NDEx account, the first time you sign in after this update, you will be asked to go through an email verification process.
While this is a simple standard procedure, we strongly recommend you execute it using the same computer and web browser to avoid unexpected behaviors. This change enhances security and unifies the sign in experience among tools within the Cytoscape ecosystem, including NDEx IQuery and the upcoming Cytoscape Web. - The X (formerly Twitter) feed widget has been removed from the NDEx home page to allow better content management. The NDEx X (formerly Twitter) account remains active and will continue to be used as an outreach channel.
- Other performance improvements and miscellaneous bug fixes.
Supported Web Browsers
We recommend that you always update your preferred web browser to the latest version whenever possible. NDEx fully supports the following browsers:
- Google Chrome
- Mozilla Firefox
While we don't discourage the use of alternative web browsers, we cannot guarantee that all features will be available and functioning as intended.
For example, if using the Brave browser, you must disable the "Brave Shield" otherwise the "Open in Cytoscape" feature in the NDEx network viewer will not work.
If you run into any issues while using NDEx, please let us know! You can easily Report a bug in NDEx using our web form.
NDEx Release Notes - Q3-2024 Update (v2.5.7)
September 1st, 2024
Overview
The Q3-2024 Update is a maintenance release and does not include any new features or API changes. Improvements in this release:
- The NDEx server has been upgraded to run Apache Tomcat 10 and Java 21.
- A new Swagger UI page for the NDEx API documentation is now available in the DEVELOPER'S section of the NDEx Documentation website.
- In the NDEx Network Viewer (NNV), the button to "Switch to Classic Mode" has been removed and the option moved to the top of the existing dropdown menu.
- Other performance improvements and miscellaneous bug fixes.
Supported Web Browsers
We recommend that you always update your preferred web browser to the latest version whenever possible. NDEx fully supports the following browsers:
- Google Chrome
- Mozilla Firefox
While we don't discourage the use of alternative web browsers, we cannot guarantee that all features will be available and functioning as intended.
For example, if using the Brave browser, you must disable the "Brave Shield" otherwise the "Open in Cytoscape" feature in the NDEx network viewer will not work.
If you run into any issues while using NDEx, please let us know! You can easily Report a bug in NDEx using our web form.
NDEx Release Notes - Q1-2024 Update (v2.5.6)
February 15th, 2024
Overview
The Q1-2024 Update is a maintenance release and does not include any new features or API changes. Improvements in this release include:
- Digital Object Identifiers (DOIs) for networks in NDEx are now automatically minted and sent to users upon request. While providing a more streamlined user experience, the removal of "manual" minting by the NDEx team means that users can no longer cancel a DOI request once it's been submitted. If users wish to modify a network after they have requested a DOI, they'll need to create a copy of their network, apply the desired changes and then submit a new DOI request.
- Minor user interface improvements (new tooltips, better error messages)
- Fixed a few bugs that caused issues when rendering networks
- Visual style improvements for CX2 networks
Content
STRING v12.0 data were added to NDEx as protein-protein interaction networks filtered according to the interaction's confidence score. Besides the full dataset (boasting almost 7 million interactions), we provide high-confidence networks filtered for scores >0.7, >0.95 and >0.99 that reduce the number of interactions to 237k, 59k and 29k respectively.
In addition, several networks generated under the Cancer Cell Map Initiative have also been added and can be used to to analyze custom gene sets via the NDEx IQuery web application.
New and existing featured content can be conveniently accessed using the "Featured Networks" dropdown menu available on the NDEx Public Server's landing page.
Supported Web Browsers
We recommend that you always update your preferred web browser to the latest version whenever possible. NDEx fully supports the following browsers:
- Google Chrome
- Mozilla Firefox
While we don't discourage the use of alternative web browsers, we cannot guarantee that all features will be available and functioning as intended.
For example, if using the Brave browser, you must disable the "Brave Shield" otherwise the "Open in Cytoscape" feature in the NDEx network viewer will not work.
If you run into any issues while using NDEx, please let us know! You can easily Report a bug in NDEx using our web form.
NDEx Release Notes - Q4-2023 Update (v2.5.5)
November 15th, 2023
Overview
The Q4-2023 Update includes new server features, improvements and minor bug fixes.
Features and Improvements
- The Cytoscape Exchange Format Version 2 (CX2) is now supported in NDEx!
While we are slowly migrating all our infrastructure toward the new CX2 format, the original CX specifications are still supported. We will begin deprecating CX once the migration is completed and NDEx 3.0 is released later in 2024.
CX2 is a major revision of the CX specifications and aims to make the data model simpler to understand and use by developers. CX2 has also a smaller memory footprint, is optimized for processing networks while streaming and makes them more compact to improve data transfer speed.
For more information and examples, please review the CX2 Documentation. - Several NDEx server changes have been implemented to support the new CX2 specifications:
- Support for the optional "type" attribute in CX2 visual style mappings.
- Disallow the declaration of alias and default values in network attributes.
- Disallow NaN and infinity as CX2 data values.
- Other improvements include but are not limited to:
- Better support of node z-location (z-index) in the network viewer.
- Improved error reporting in the CX2 validator
Content
New and existing featured content can be conveniently accessed using the "Featured Networks" dropdown menu available on the NDEx Public Server's landing page.
Supported Web Browsers
We recommend that you always update your preferred web browser to the latest version whenever possible. NDEx fully supports the following browsers:
- Google Chrome
- Mozilla Firefox
While we don't discourage the use of alternative web browsers, we cannot guarantee that all features will be available and functioning as intended.
For example, if using the Brave browser, you must disable the "Brave Shield" otherwise the "Open in Cytoscape" feature in the NDEx network viewer will not work.
If you run into any issues while using NDEx, please let us know! You can easily Report a bug in NDEx using our web form.
NDEx Release Notes - Q3-2023 Update (v2.5.4)
August 17st, 2023
Overview
The Q3-2023 Update is a maintenance release. It only includes a new implementation of the Google Oauth2 sign in feature. This release doesn't include any new features, API changes or bug fixes.
Content
As usual, all new and existing featured content can be conveniently accessed using the "Featured Networks" dropdown menu available on the NDEx Public Server's landing page.
Supported Web Browsers
We recommend that you always update your preferred web browser to the latest version whenever possible. NDEx fully supports the following browsers:
- Google Chrome
- Mozilla Firefox
While we don't discourage the use of alternative web browsers, we cannot guarantee that all features will be available and functioning as intended. If you run into problems using a browser other than those specified above, please let us know! You can easily Report a bug in NDEx using our web form.
NDEx Release Notes - Q3-2022 Update (v2.5.3)
August 1st, 2022
Overview
The Q3-2022 Update is a maintenance release. It includes bug fixes and server performance improvements. This release doesn't include any new features or API changes.
Content
- The Human Protein Atlas aims to map all the human proteins in cells, tissues, and organs using an integration of various omics technologies, including antibody-based imaging, mass spectrometry-based proteomics, transcriptomics, and systems biology. Selected data from The Human Protein Atlas v21.0 are now available in NDEx.
- HumanNet provides access to high-quality integrated functional protein networks. With its recent new release, HumanNet v3 provides >99% genome coverage.
As usual, all new and existing featured content can be conveniently accessed using the "Featured Networks" dropdown menu available on the NDEx Public Server's landing page.
Supported Web Browsers
We recommend that you always update your preferred web browser to the latest version whenever possible. NDEx fully supports the following browsers:
- Google Chrome
- Mozilla Firefox
While we don't discourage the use of alternative web browsers, we cannot guarantee that all features will be available and functioning as intended. If you run into problems using a browser other than those specified above, please let us know! You can easily Report a bug in NDEx using our web form.
NDEx Release Notes - Q4-2021 Update (v2.5.2)
October 19th, 2021
Overview
The Q4-2021 Update includes new features and improvements both in the user interface and the server back-end. Please review this document carefully prior to using NDEx v2.5.2.
New Features and improvements
- Cytoscape 3.9 New Features are now supported in NDEx; these include "Table visual styles" as well as a number of new "Network visual styles" such as node/edge label rotation, haystack edges and edge Z order. If you use Cytoscape and NDEx for your work, we strongly recommend you upgrade to Cytoscape 3.9 to capitalize on all newly supported features.
- The Request DOI feature has been streamlined and semi-automated to provide faster turn-around for DOI record assignment. These changes are invisible to users, neither the interface nor the user experience have been modified. In addition, all DOIs will automatically point to the New Network Viewer (NNV) page rather than the classic viewer; if users want the DOI record to point to the classic viewer, they should submit their DOI request normally and then Contact Us to specifically request their DOI record be pointed to the classic viewer page.
- The default sorting criteria in "Network Sets" has been changed for consistency: networks are now sorted by Last modified throughout the entire NDEx web application user interface.
- The New Network Viewer (NNV) now allows multiple selection of nodes/edges up to a maximum of 10,000 elements for any network displayed in NDEx.
- Node and Edge tables in NNV now support pagination and deliver improved performance and usablity.
Content
- FunCoup is one of the most comprehensive functional association networks of genes/proteins available. FunCoup 5 now includes 22 species from all domains of life, and the source data for evidences, gold standards, and genomes have been updated to the latest available versions. This network has over 5 million edges and cannot be displayed fully in NDEx. You can "Switch to Classic Mode" to have the legacy viewer display a random sample; alternatively, you can query the full network, save the result in NDEx and/or import it in Cytoscape for visualization and further analysis.
As usual, all new and existing featured content can be conveniently accessed using the "Featured Networks" dropdown menu available on the NDEx Public Server's landing page.
Supported Web Browsers
We recommend that you always update your preferred web browser to the latest version whenever possible. NDEx fully supports the following browsers:
- Google Chrome
- Mozilla Firefox
While we don't discourage the use of alternative web browsers, we cannot guarantee that all features will be available and functioning as intended. If you run into problems using a browser other than those specified above, please let us know! You can easily Report a bug in NDEx using our web form.
NDEx Release Notes - Q2-2021 Update (v2.5.1)
May 28th, 2021
Overview
The Q2-2021 Update is a major release that includes new features and improvements both in the user interface and on the server side. Please review this document carefully prior to using NDEx v2.5.1.
New Features and improvements
- Node and Edge tables have been revamped and are now displayed in their own tabs in the NNV info panel. The tables are structured similarly to the node and edge tables in the Cytoscape desktop application to guarantee a consistent user experience. The tables are only used to display the data when multiple graphic elements are selected. For individual elements, the existing pop up inspector window is used. Please note: for very large networks, multiple selection and the new Nodes/Edges tables are not enabled yet.
- The new Query External Database feature allows users to send a query to an external database resource using the gene names of any network in NDEx. Users can choose to send the query using either all or a selection of the genes in the network. Currently, the 2 available external database options are MSigDB and IQuery and more options will be added in future releases.
- It is now possible to Share Network, Request DOI and Cancel DOI Request directly in NNV without having to first Switch to Classic Mode. Additional functions (such as Clone Network) will be ported over to NNV in future releases.
- When viewing large networks in NNV, a new Fit Content button now allows users to reset the view and display the entire network.
- The Neighborhood Query has been improved to search on all available node text fields by default; with this improvement, querying a network for 1 (or more) genes, will always return a result, even if the gene names are stored under an arbitrary node property instead of the "name" property. You can also query a network for a Uniprot ID (i.e. Q08477) and the query result will return the neighborhood of the node with that ID stored under one of its properties.
- Performance improvements provide faster access to networks when using a Sharable URL and you will find better tooltips and user-friendly error messages throught the entire NDEx user interface.
- Finally, NDEx users can now receive Usage Statistics for their published networks. These are sent by e-mail on a monthly basis and users can decide to opt-out using the control box available in their Account Settings.
Content
- The new NDEx Cancer Collection gathers both cancer-focused network models and other networks relevant to cancer research. The networks come from many sources and each retains its original visual style, naming conventions, and data schema. Many of the networks contain the reference of the original publication in which they are described. Occasionally there may be differences between the appearance of the network in NDEx and the corresponding figure in the publication.
- The Causal Biological Networks (CBN) database is composed of multiple versions of over 120 modular, manually curated, BEL-scripted biological network models supported by over 80,000 unique pieces of evidence from the scientific literature. They represent causal signaling pathways across a wide range of biological processes including cell fate, cell stress, cell proliferation, inflammation, tissue repair and angiogenesis in the pulmonary and vascular systems. The CBN networks in NDEx are only human pathways; mouse and rat pathways are available for download on the CBN website.
- BioPLEX3 includes nearly 120,000 interactions among nearly 15,000 proteins and is the most comprehensive experimentally derived model of the human interactome to date. Because each protein’s network position reflects its subcellular localization, biological function, and disease association, these networks have been powerful tools for the study of thousands of uncharacterized proteins. They have also provided insights into interactome modularity and organization.
As usual, all new and existing featured content can be conveniently accessed using the "Featured Networks" dropdown menu available on the NDEx Public Server's landing page.
Supported Web Browsers
We recommend that you always update your preferred web browser to the latest version whenever possible. NDEx fully supports the following browsers:
- Google Chrome
- Mozilla Firefox
While we don't discourage the use of alternative web browsers, we cannot guarantee that all features will be available and functioning as intended. If you run into problems using a browser other than those specified above, please let us know! You can easily Report a bug in NDEx using our web form.
NDEx Release Notes - Q4-2020 Update (v2.5.0)
November 5th, 2020
Overview
The Q4-2020 Update is a major release that includes a brand new network viewer as well as performance improvements. The Q4 2020 Update doesn't include any public API changes.
New Features
This section provides information about new features and improvements introduced with the Q4-2020 Update. Please review this document carefully prior to using NDEx 2.5.0 for your work.
New Network Viewer (NNV)
NNV is capable of rendering very large networks with up-to 2 million objects thanks to the implementation of a brand new rendering engine. While NNV can faithfully display small networks, large networks are displayed using a "simplified style" that makes possible handling millions of objects within the limitations of an individual browser's tab.
NNV boasts a new node/edge inspector that can be triggered by clicking a node/edge in the graphic display. The info panel on the right hand side can be resized or completely collapsed to maximize the screen space used for network visualization.
Neighborhood Query results are now displayed in a "split view" fashion, thus providing both the fine detail of the subnetwork result and the overall view of the complete starting network.
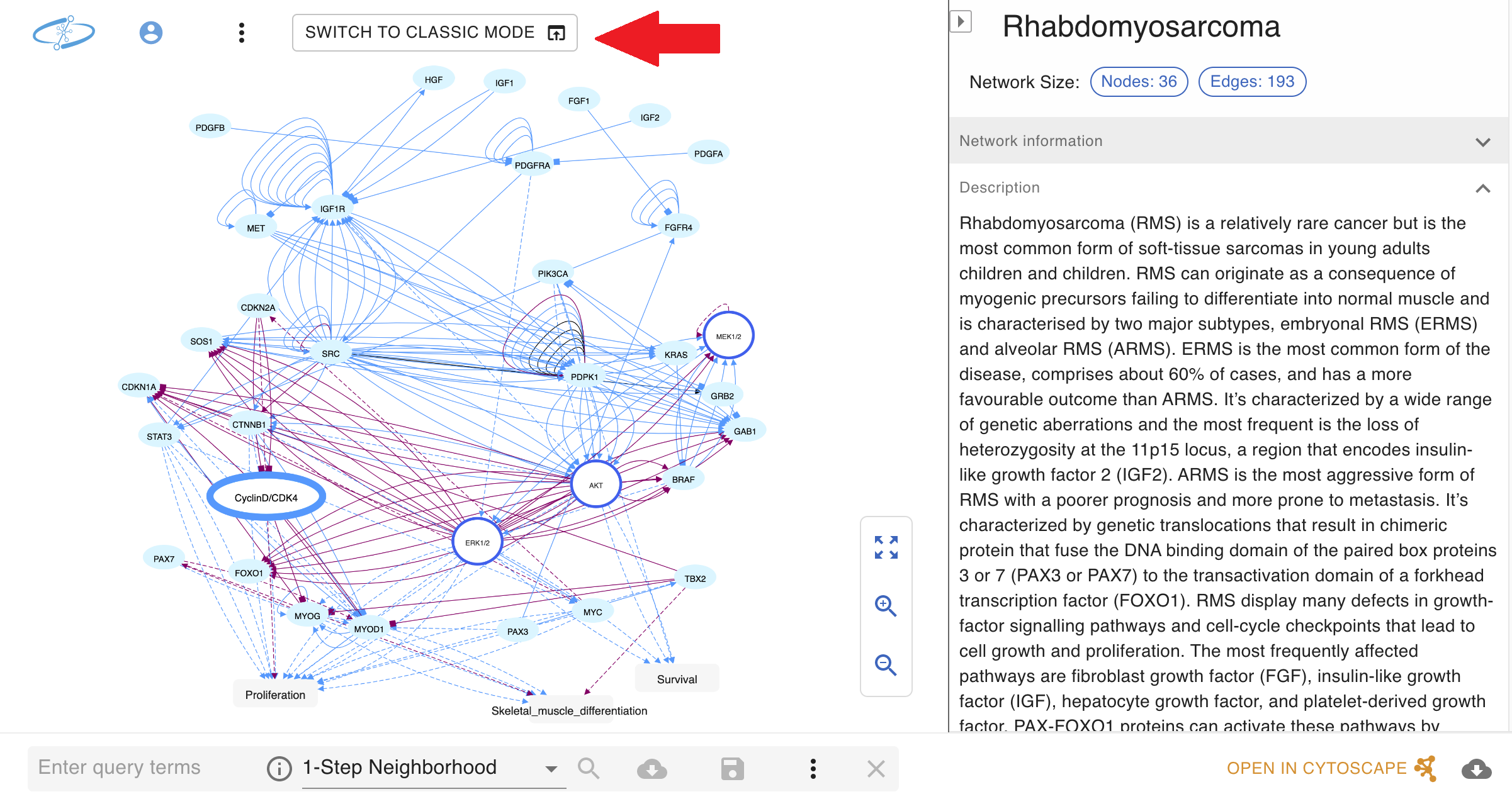
!!! IMPORTANT NOTE: as NNV is still in active development, not all existing NDEx features have been ported over yet. Users can still access all existing NDEx features by switching back to "Classic Mode" with the button in the top left corner of the NNV page (red arrow in screenshot below).
New server-side Network validator
The CX validator has been refactored and improved. Existing networks with visual styling issues are now fixed. Specifics about the server-side improvements can be provided upon request.
Content
- The new NDEx Coronavirus Collection contains data from know repositories and published research articles. So far, the collection counts 56 networks including interaction data from BioGRID, Signor, IMEx/IntAct as well as a number of quality networks from hi-profile peer-revied publications. This Collection is curated, maintained and updated regularly by the NDEx Team.
- The Cancer Center for Systems Biology at Dana Farber Cancer Institute has published the Human Reference Protein Interactome Mapping Project (HuRI), a collection of 49 high-quality, cancer-focused networks.
All new and existing featured content can be conveniently accessed using the "Featured Networks" dropdown menu on the NDEx Public Server's landing page
Supported Web Browsers
We recommend that you always update your preferred web browser to the latest version whenever possible. NDEx fully supports the following browsers:
- Google Chrome
- Mozilla Firefox
- Full network display threshold is increased from 12000 edges to 20000 edges for networks with layouts.
- Bug fix: DOI request where causing an error when networks have ‘$’ or ‘\’ in the network description field.
- Bug fix: Setting edge visibility in Cytoscape was not handled properly when rendering the network in the NDEx web app.
- Google Chrome
- Mozilla Firefox
- Safari
- The cytoscape.js library was updated to v3.12.1 for enhanced graphical rendering.
- Added support for the Node Label Justification property.
- Improvement of the Network Tabular View page that now correctly handles columns with no values in the cells.
- Google Chrome
- Mozilla Firefox
- Safari
- The search engine was tuned to assign more weight to matched search terms found in "Network Name" and "Description" fields.
- The Solr engine was upgraded to v8.1.1 and adjustments applied to the schema to boost performance and quality of the results.
- Optimization of the Query engine to improve performance for "Interconnect" queries.
- REST server: fixed a regression bug in the "update aspects" function.
- Query service: fixed bugs that caused an incorrect edge count and missing edges in the Interconnect Query method.
- Web UI:
- Bug fix: certain node/edge attributes were not displayed in table view.
- Bug fix: sorting functions not behaving correctly in table view.
- Various improvements in the network displayer.
- Improvements in the Featured content and Featured Networks section on the NDEx Public Server's landing page.
- Improvement in the CX validator. The server now returns an error if the CX network is missing the "node aspect". It also checks for dangling IDs in the "cyVisualProperties" and "cartesianLayout" aspects.
- Fixed a bug where the "USER_STORAGE-LIMIT" parameter was not read correctly by the server.
- The server no longer returns an exception when users click the account verification link more than once.
- Fixed a bug where the edge count was wrong in the Interconnect query.
- Auto-closae timers have been removed for bulk network operations.
- Cytoscape Annotations are now supported and can be faithfully displayed in the NDEx network visualizer.
- The checkbox option to enable "Full Indexing" of a network is now disabled if a user is not the owner of that network.
- Deprecating @context aspect. Use of ‘@context’ network attribute is recommended.
- Deprecating the Numeric Range Precondition Element in CX format.
- Deprecating the Provenance aspect.
- The provenance tab was removed from the network view page in the NDEx web UI to simplify provenance handling.
- The NDEx server no longer generates the ndexStatus aspect in networks.
- Run a neighborhood query on a large network and use the "Set Sample" button to set the result to be the displayed sample.
- Click the "More" button in the network view page and choose "Set Network Sample" to specify the UUID of another network to use as sample
- Interconnect Query
A new query option, “Interconnect”, has been added to the query menu on network pages. An Interconnect returns only short paths between the nodes selected by the query string. The resulting subnetwork attempts to answer the question “How are these related to each other?”. It therefore is only appropriate for queries selecting 2 or more nodes. Query nodes for which no short paths are found to any other query node will appear as “orphans”, nodes that have no edges. - Improved Performance
The Neighborhood Query now supports networks of any size and always returns a result up to 50,000 edges. Smaller query results will be presented in graphic form while larger results will only be shown in tabular form to avoid long delays. - Autosave Feature
If the query result is larger than 50,000 thus exceeding the web browser's capability, the new Autosave feature will offer to automatically save the query result to the user's account. This feature is only available to logged in users; anonymous users will be prompted to log in and will need to re-run the query in order to have the result automatically saved. - Download Result
Query results up to 50,000 edges can now be downloaded as tab-delimited text files (TSV). This feature is available both to anonymous and logged-in users and can be accessed using the new "Download Result" button available in the query result page. - updated our network visualizer to use the latest cytoscape js version available (v3.2.9)
- fine-tuned its parameter to obtain a fast and smooth graphic rendering
- changed sampling logic; when a network exported from Cytoscape is saved in NDEx, our visualizer now displays it in full if the network has up to 12,000 edges! For larger networks, a 300 edges sample view is generated instead.
- Open in Cytoscape - Beginning with v2.2.1, users can open an NDEx network in Cytoscape directly from the NDEx web User Interface. This feature is triggered by a new "Cytoscape button" available in the network view page. Due to technical limitations, right now this feature can only be used if the following conditions are met:
- Cytoscape 3.6.0 or higher must be installed and running on your computer and have the CyNDEx-2 app installed
*** Note: starting with Cytoscape 3.6.1, CyNDEx-2 will become a pre-installed, core app. *** - The network to be opened in Cytoscape must be either Public or Private and have the Sharable URL enabled
- Cytoscape 3.6.0 or higher must be installed and running on your computer and have the CyNDEx-2 app installed
- @context Editor - NDEx 2.2.1 brings back the possibility to control the namespaces associated to any network. The @context Editor can be accessed from the network page and dinamically adapts to the user's privilege level for that specific network. Namespaces can always be viewed but can be added or modified only if the user has "Edit" or "Admin" privileges on the network. Defining namespaces using the @context Editor (or other method) is important as it will make possible for the NDEx UI to use the identifiers specified in the network (represents, alias and other network attributes) and automatically generate URL to point to relevant external resources. Defining namespaces for a network is not mandatory but highly recommended.
- Expanded Network Indexing Policy - In NDEx v2.2.1, the SOLR indexing policy has been modified to optimize server performance, relevance of search results and system scalability. There are now 3 tiers of indexing complexity the users can choose from:
- Tier 0 (T0)
- This is the default indexing behavior and applies to all PRIVATE and PUBLIC networks written to NDEx regardless of their origin (file loading, script, cytoscape)
- T0 corresponds to the PRIVATE (Not Searchable) or PUBLIC (Not Searchable) visibility options currently available in the NDEx UI
- Under T0, the only parameter that is indexed is the network's UUID
- Tier 1 (T1)
- This is the extended indexing behavior and can be applied to any PRIVATE or PUBLIC networks already existing in NDEx
- T1 corresponds to the PRIVATE or PUBLIC visibility options currently available in the NDEx UI
- T1 can only be selected manually in the NDEx UI, either on a per-network basis or as a bulk action
- Under T1, the following network attributes are indexed for search: UUID, name, description, reference, disease, author, labels, tissue, methods, rightsHolder, rights, organism, networkType
- Under T1, a new add-on option will become available via a selectable checkbox in the network property editor to optionally add "full node indexing". If selected, the network will be indexed according to the Tier 2 specs listed below.
- Tier 2 (T2)
- This is the most comprehensive indexing tier
- Full indexing is only available on a per-network basis via a selectable checkbox in the network property editor; this option is not available as a bulk action
- By default, the "Full Index" checkbox is NOT selected
- When selected, in addition to the network attributes indexed under T1, the following node attributes will also be indexed for search: nodeName, represents, alias
- This feature uses a lot of resources and should be used sparingly
- "Certified" and "Pre-certified" networks are automatically indexed under T2
- CyNDEx v4.0.3
This is the original NDEx Cytoscape app and can be used with Cytoscape v3.3 or higher. There are no new features in this version of the CynDEx app; click the version number to access the app's store page and review the details about bug fixes and other improvements.
If you need help with the CyNDEx app, please review our updated CyNDEx Tutorial. - CyNDEx-2 v2.2.1
CyNDEx-2 is the latest NDEx Cytoscape app and can only be used with Cytoscape v3.6 or higher. CyNDEx-2 relies on the REACT framework, uses JX Browser and will become a "core app" in the upcoming Cytoscape v3.6.1 release.
CyNDEx-2 has an improved user interface that makes it easier to browse, import, annotate and export networks. In addition, it makes possible to use the new "open in Cytoscape" feature described above in this document.
If you work with Cytoscape v3.6 or higher, we recommend using CyNDEx-2 to take full advantage of allsnew features and improvements.
For instructions about the app and its use, click the version number above to access the app's store page.
While we don't discourage the use of alternative web browsers, we cannot guarantee that all features will be available and functioning as intended. If you run into problems using a browser other than those specified above, please let us know! You can easily Report a bug in NDEx using our web form.
NDEx Release Notes - Q2-2020 Update (v2.4.5)
April 1st, 2020
Overview
The Q2-2020 Update is a maintenance release that includes bug fixes and performance improvements. The Q2 2020 Update doesn't introduce any new features or API changes.
Improvements and bug fixes
Content
A new host-pathogen map and relevant drug-target interactions for SARS-CoV-2 are now publicly available.
Data from Inoue et al. (2019) on ligand-mediated GPCR G-protein-coupling for 148 GPCRs has also been added to the pool of publicly available networks in NDEx.
All new and existing featured content can be conveniently accessed using the "Featured Networks" dropdown menu on the NDEx Public Server's landing page
Supported Web Browsers
We recommend that you always update your preferred web browser to the latest version whenever possible. NDEx fully supports the following browsers:
While we don't discourage the use of alternative web browsers, we cannot guarantee that all features will be available and functioning as intended.
NDEx Release Notes - Q1-2020 Update (v2.4.4)
January 15th, 2020
Overview
The Q1-2020 Update is a maintenance release that includes performance and stability improvements and several bug fixes, both in the REST server and WebApp codebases. The Q1-2020 Update doesn't introduce any new features or API changes.
Web UI Improvements
This section includes a brief overview of some user interface improvements included in the NDEx v2.4.4 release.
For more details and a full list of changes, please review the following pages:
Content
The latest GeneHancer data from the GeneCards Suite is now available in our Featured Networks collection.Other existing content was also updated using the latest data available.
Supported Web Browsers
We recommend that you always update your preferred web browser to the latest version whenever possible. NDEx fully supports the following browsers:
While we don't discourage the use of alternative web browsers, we cannot guarantee that all features will be available and functioning as intended.
NDEx Release Notes - Q3-2019 Update (v2.4.3)
August 26th, 2019
Overview
The Q3-2019 Update is a minor release including bug fixes and new features for the Search and Neighborhood Query functions.
New Features
This section provides information about new features and improvements introduced with the Q4-2019 Update. Please review this document carefully prior to using NDEx 2.4.3 for your work.
Attribute Search and Query - Both the Search and Query functions were extended to search the "member" attribute for node types "proteinfamily" and "complex". This new features allows to find nodes that represent molecular complexes or generic protein families based on the participating genes specified under the "member" node attribute.
Query Node Highlight - When performing a neighborhood query, the NDEx UI can now highlight all query terms in the graphic rendering thus allowing immediate identification of the query nodes. This new feature works in parallel with the other new feature described above, by higlghlighting a "proteinfamily" or "complex" node that contain one or more query terms. A new button allows to toggle Query Node Highlight on and off. By default, this new feature is turned on.
Improvements
This section includes a brief overview of the improvements included in the NDEx v2.4.3 release:
Content
Several networks available in the Featured Networks dropdown menu in the NDEx Search box were updated using the latest data available. These include but are not limited to: STRING, BioGRID and Signor.
Supported Web Browsers
We recommend that you always update your preferred web browser to the latest version whenever possible. NDEx fully supports the following browsers:
– Google Chrome
– Mozilla Firefox
– Safari
While we don't discourage the use of alternative web browsers, we cannot guarantee that all features will be available and functioning as intended.
NDEx Release Notes - Q2-2019 Update (v2.4.2)
June 7th, 2019
Overview
The Q2-2019 Update is a maintenance release and only includes bug fixes and other minor system system improvements. No new features have been added to the NDEx server, API and web application UI.
Improvements and bug fixes
This section includes a brief overview of some of the improvements included in the NDEx v2.4.2 release:
Content
New content available in NDEx includes the Human Protein Atlas (HPA), the Atlas of Cell Signaling Network (ACSN v2.0) and ProteomeHD. Existing resources have been updated with the latest data available, such as STRING v11.0. Use the Featured Networks dropdown menu in the NDEx Search box to access these new networks and many more.
Supported Web Browsers
We recommend that you always update your preferred web browser to the latest version whenever possible. NDEx fully supports the following browsers:
– Google Chrome
– Mozilla Firefox
– Safari
While we don't discourage the use of alternative web browsers, we cannot guarantee that all features will be available and functioning as intended.
NDEx Release Notes - Q1-2019 Update (v2.4.1)
April 26th, 2019
Overview
The Q1-2019 Update includes performance improvements, bug fixes and other minor system tweaks. No new features have been added to the NDEx server, API and web application UI.
Improvements
This section includes a brief overview of some of the improvements included in the NDEx v2.4.1 release:
REST Server
Neighborhood Query
Web Application/UI
Content
Thanks to the latest improvements and a strong collaborative effort with the WikiPathways team, over 500 high quality, manually curated, human pathways are now publicly available in NDEx. Please search for WikiPathways in NDEx or CLICK HERE to access this rich new source of content.
In addition, severasl other existing network collections have been updated with the latest data available: BioGRID, STRING, BindingDB, Drugbank, CTD and more. You can easily explore all of them using the Featured Networks button available in the NDEx Search Box.
Supported Web Browsers
We recommend that you always update your preferred web browser to the latest version whenever possible. NDEx fully supports the following browsers:
– Google Chrome
– Mozilla Firefox
– Safari
While we don't discourage the use of alternative web browsers, we cannot guarantee that all features will be available and functioning as intended.
NDEx Release Notes - Q4-2018 Update (v2.4.0)
December 20th, 2018
Overview
The Q4-2018 Update includes new features and improvements in the NDEx server, API, web application and documentation. All features in NDEx v2.4.0 are deployed and available on the NDEx Public Server.
New Features and Improvements
This section provides information about new features and improvements introduced with the Q4-2018 Update. Please review this document carefully prior to using NDEx 2.4.0 for your work.
New Landing Page - The NDEx landing page was completely redesigned and now provides more information about NDEx and how scientist can use it in their everyday research. The NDEx Twitter Feed keeps you informed on interesting topics related to network biology and the Featured Content panel provides quick access to network collections, new publications and upcoming conferences. The Top Menu bar grants easy access to our documentation, contact and Bug Report forms, while up-to-date Release Notes can be accessed by clicking the version number right next to the NDEx logo.Note: if you encounter any page visualization issues, please clear your browser's cache and reload the page.
Google Sign Up - Creating an account in NDEx 2.4.0 is easier than ever; users have now the option to Register with Google, provided that they already have an active Google account. This new feature makes creating an account as easy as 2 clicks and will associate your Gmail address to your account, so you'll also be able to seamlessly Log in to NDEx with Google in the future. After you succesfully Sign Up with Google, our system will send you an email with your log in credentials that you can use if you are interested in using your NDEX account programmatically via custom scripts or applications. For additional details and step by step instructions with screenshots, please refer to the user manual Creating and Using an NDEx Account.
Neighborhood Query - In NDEx 2.4.0, the Neighborhood Query feature was improved and simplified. Users can now choose among 4 different types of query using a dropup menu: Neighborhood, Adjacent, Direct and Interconnect. In addition, when performing a Neighborhood or Adjacent query, the "Depth" can also be specified. Embedded documentation explains in detail the available query types and can be accessed by clicking the small "info" icon next to the query type drop up menu. Additional info and screenshots can be found in the manual on Finding and Querying Networks in NDEx.
Note: the Advanced Query feature is still in development and will be available later in 2019.
GraphML Exporter - In addition to exporting networks as GSEA Gene Sets, NDEx 2.4.0 also lets users export networks as GraphML objects. To export a network, click the More button in the network view page and select "Export" in the dropup menu; then choose the export format you want and click the Export Network button. Your export request will be qued as a task by the NDEx server. To check its status and download the exported file, please visit the "My Tasks & Notifications" tab in your NDEx account page.
Additional exporters are currently under development and will become available in 2019. These include an Excel Spreadsheet exporter, a tabular file exporter (csv and Tab-delimited text) and a CytoscapeJS exporter.
Cancel DOI Request - The request DOI feature was improved by adding the option to Cancel a DOI Request once it has been submitted. This improvement is particularly useful when users realize they have not included all information in the network before submitting their request. Cancelling a submitted DOI request makes the network editable again so that missing/incorrect information can be modified. If a DOI request is cancelled, users must submit a new request if they want a DOI assigned to their networks. For more information, please review the manual on Publishing in NDEx.
Account Management - Improvements in the account management logic now allow users to reset the password of their NDEx account using either their account name or e-mail address. In addition, whenever users change the email address associated to their account, the systems sends an email notification to the old address, just in case the change is unauthorized or executed by mistake.
CX Optimization - The CX format was simplified and made more efficient through a number of changes that are described in more detail in the CX Data Model document. The changes in NDEx v2.4.0 include but are not limited to:
cx2js Library - The code that managed CX JSON to Cytoscape.js JSON conversion was refactored into a general purpose npm package, allowing its re-use in other projects. Example use cases were included for package consumers, and unit tests were introduced to increase reliability and prevent regression. The font handling of the library was updated, fixing several bugs, including the handling of Java Logical Fonts and label positions. The cx2js GitHub repo is available HERE.
Improved Documentation - The documentation for NDEx v2.4.0 was revised and improved. All manuals are brand new and include up-to-date screenshots of the v2.4.0 web UI. All technical docs were updated to reflect the latest changes in the data model, CX format and API functions. A new Developing in NDEx - README!!! page was created to centralize all developer's related info and links to code repositories. Finally, more embedded documentation was added to the web UI in the form of tooltips and info modal windows. If you are new to NDEx, we recommend you beging with the updated Quick Start Guide and then explore the other manuals and documents using the blue Doc Navigation Menu.
Cytoscape Apps
The integration between NDEx and Cytoscape keeps improving and the CyNDEx-2 module is a core component in Cytoscape 3.7. We strongly recommend you download the latest Cytoscape version available and then update the CyNDEx-2 core component directly from the Cytoscape App manager if necessary.
All users relying on older Cytoscape versions can still take full advantage of the NDEx-Cytoscape integration thanks to the CyNDEx (v4.0.3) app! This is the first, original NDEx Cytoscape app and can be used with Cytoscape v3.3 or higher. There are no new features in this version of the CynDEx app; click the version number to access the app's store page and review the details about bug fixes and other improvements.
If you need help with the CyNDEx app, please review our updated CyNDEx Tutorial.
Supported Web Browsers
We recommend that you always update your preferred web browser to the latest version whenever possible. NDEx fully supports the following browsers:
– Google Chrome
– Mozilla Firefox
– Safari
While we don't discourage the use of alternative web browsers, we cannot guarantee that all features will be available and functioning as intended.
NDEx Release Notes - Q3-2018 Update (v2.3.1)
July 25th, 2018
Overview
The Q3-2018 Update brings new features to the NDEx web application and improvements in Cytoscape integration. All features in NDEx v2.3.1 are deployed and available on the NDEx Public Server.
NDEx v2.3.1 is also available as an installation bundle for deploying private instances of the NDEx server. For more information and to download, please refere to the NDEx Installation Instructions.
New Features and Web User Interface Improvements
This section provides information about new features and improvements introduced with the Q3-2018 Update. Please review this document carefully prior to using NDEx 2.3.1 for your work.
Export Query Result - In NDEx 2.3.1, Neighborhood Query results can be exported in tabular format (tab-delimited text) directly by using the new Download Result button available in the Query result page. This new feature is available to all users, both anonymous and logged-in. For a detailed guide on using the Neighborhood Query, please refer to the user manual Finding and Querying Networks in NDEx.Note: the Advanced Query feature is still in development and will be available later in 2018.
Google Sign-In for Safari - Existing initialization issues have been resolved and the convenient Google authentication feature can now be used with the Safari web browser.
Partial Support for Cytoscape Collections - NDEx v2.3.1 allows users to set the "Read-only" flag for Cytoscape collections either as a bulk action or directly in the network view page.
In addition, users can now also change the "Visibility" of a Cytoscape collection; however, at this time the feature is only available as a bulk action from your "My Account" page.
Display Networks in External Applications - This feature is tailored to developers and allows networks to be displayed in any application via the NDEx viewer and seamlessly opened in Cytoscape if desired, regardless whether networks are stored in NDEx or not. This feature is currently being used by some of our collaborators within the ITCR projects portfolio. If you are interested in displaying networks in your application and don't want to re-invent the wheel, Contact Us and we'll be happy to help!
Open in Cytoscape - The existing feature has been improved as part of the ongoing effort to better integrate NDEx and Cytoscape. Private networks can now be opened in Cytoscape along with unsaved "Neighborhood Query" resultsNote: in order to use this feature, Cytoscape 3.6.0 or later must be running on your machine, the latest CyNDEx-2 App (v2.2.3) installed and the CyREST service listening on its default port (1234).
Network Table Customization - The network table in your "My Account" page has been improved to preserve user-defined sorting, filtering, multiple selection, and paging. Navigation has also been improved: users can now use the browser Back and Forward buttons to navigate to previously-visited network pages.
Visual Style Mappings - The NDEx network viewer has been improved to better translate visual style mappings between the Cytoscape desktop application and Cytoscape.js on the web. Pie Charts created in Cytoscape can now be rendered in NDEx; in addition, the following mappings are also rendered correctly in v2.3.1: Node Border Line Types, Node Label Position, Node Shape, Edge Line Type, Edge Source/Target Arrow Shape, Node and Edge Label Font Face, Node Label Position.
Remove Shared Networks - In NDExv2.3.1, users can reduce clutter in their accounts by removing networks that other users have previously shared with them. The "Remove From My Networks" feature is available both for individual networks in the network view page (click "More" and choose "Remove From My Networks") or as a bulk action directly in your "My Account" page.
Custom Network Sample - For large networks, the NDEx network viewer displays a randomly generated 300 edges sample to present a visually interpretable subset of the data. In NDEx v2.3.1, users have two methods to specify a custom sample to be displayed therefore improving the presentation of a large network:
For more details, please review the user manual Finding and Querying Networks in NDEx.
Cytoscape Apps
The integration between NDEx and Cytoscape keeps improving: the CyNDEx-2 (v2.2.3) app has just been released that includes bug fixes and new features as already described earlier in this document. We strongly recommend users download the latest Cytoscape version available and then update the CyNDEx-2 core component directly from the Cytoscape App manager.
All users relying on older Cytoscape versions can still take full advantage of the NDEx-Cytoscape integration thanks to the CyNDEx (v4.0.3) app! This is the first, original NDEx Cytoscape app and can be used with Cytoscape v3.3 or higher. There are no new features in this version of the CynDEx app; click the version number to access the app's store page and review the details about bug fixes and other improvements.
If you need help with the CyNDEx app, please review our updated CyNDEx Tutorial.
Supported Web Browsers
We recommend that you always update your preferred web browser to the latest version whenever possible. NDEx fully supports the following browsers:
– Google Chrome
– Mozilla Firefox
– Safari
While we don't discourage the use of alternative web browsers, we cannot guarantee that all features will be available and functioning as intended.
NDEx Release Notes - Q2-2018 Update (v2.3.0)
April 5th, 2018
Overview
The NDEx Q2-2018 Update provides major improvements to the end-user experience while optimizing performance and scalability. All features in NDEx v2.3.0 are deployed and available only on the NDEx Public Server. No new installation bundle will be released in Q2-2018.
New Features
This section provides information about the new features introduced with the Q2-2018 Update. Please review this document carefully prior to using NDEx 2.3.0 for your work.
Note: the Advanced Query feature has been temporarily disabled to allow improving its performance, scalability and user experience. The new Advanced Query feature will be available later in 2018.Details about new features and improvements in the Neighborhood Query are listed below and additional info and screenshots are available in the updated guide focused on Finding and Querying Networks in NDEx.
Web User Interface Improvements
In NDEx 2.3.0, we have improved network visualization, especially for all those networks that have been styled and layed out using Cytoscape. To achieve this, we have:
Cytoscape Apps
The connection between NDEx and Cytoscape has now reached full integration: the CyNDEx-2 (v2.2.2) app has become a "core" Cytoscape component in the recently released Cytoscape v3.6.1. An NDEx sample session is also available to get new users started immediately with NDEx. Click the link above to access the app's store page for a more info and a brief tutorial. Additional information can be found in the Cytoscape online manual.
All users relying on older Cytoscape versions can still take full advantage of the NDEx-Cytoscape integration thanks to the CyNDEx (v4.0.3) app! This is the first, original NDEx Cytoscape app and can be used with Cytoscape v3.3 or higher. There are no new features in this version of the CynDEx app; click the version number to access the app's store page and review the details about bug fixes and other improvements.
If you need help with the CyNDEx app, please review our updated CyNDEx Tutorial.
Technical Documentation
New features and other improvements in NDEx 2.3.0 required changes in the REST API; please review carefully the NDEx API Documentation prior to using it for your work.
Network Content
NDEx 2.3.0 brings plenty of new, high quality up-to-date content for users to browse, download and re-use. We now have automated pipelines to regularly update all manually curated pathways available in the SIGNOR Database; also available are protein-protein interaction networks for several species, generated from the BioGRID and STRING current release data.
NDEx Release Notes - Q1-2018 Update (v2.2.1)
February 1st, 2018
Overview
The NDEx Q1-2018 Update brings changes and new features to improve the end-user experience while optimizing performance and scalability. All new features and improvements in NDEx v2.2.1 are deployed and available only on the NDEx Public Server. No new installation bundle will be released in Q1-2018.
New Features
This section provides information about the new features introduced with the Q1-2018 Update. Please review this document carefully prior to using NDEx 2.2.1 for your work.
Web User Interface Improvements
In NDEx 2.2.1, the web UI includes several improvements to the general look and feel; the landing page has been decluttered to focus on the most important components: the search bar and featured collections. In addition, the landing page also shows the current NDEx version and provides direct access to the latest Release Notes.
The network view page has new embedded documentation in the form of tooltips that can be activated by hovering on buttons and other UI elements. Tooltips also provide explanatory messages in cases where a certain function is not available to the user.
Finally, the network viewer has been tweaked to render Cytoscape visual styles with higher fidelity when using discrete mappings, continous mappings and features like node and edge transparency.
Cytoscape Apps
The tight connection between NDEx and Cytoscape is now stronger than ever: with NDEx 2.2.1, user can choose between 2 different apps to round trip networks between NDEx and Cytoscape.
NOTE: all Cytoscape collections are displayed in the NDEx UI with the a black Cytoscape icon. Any single-subnetwork
collection can be converted to single network simply by importing it into Cytoscape with CyNDEx-2 and re-exporting it to
NDEx as network; we also recommend you uncheck the option to "Save as New Network" in order to update the existing copy of the network.
Technical Documentation
NDEx 2.2.1 brings a number of important back-end changes that are documented in our Technical Literature. Please review the Network Data Model document for updates in the CX specification and server changes.
New features and other improvements in NDEx 2.2.1 also required changes in the REST API; please review carefully the NDEx API Documentation prior to using it for your work.